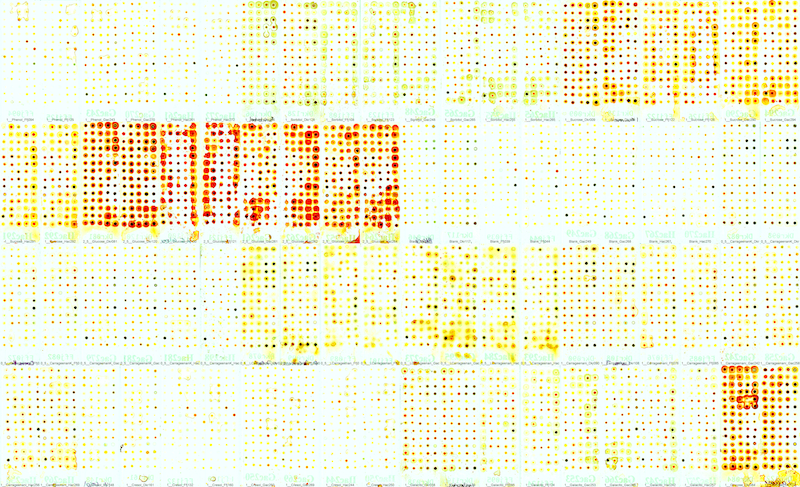Can OpsisDx distinguish between similar molecules? Let’s say between different salts or isomers of carrageenan or phenol vs cresol?
YES … we observe distinct NuTec profiles for each molecule or mixture, even for molecules with similar structures.
This provides a framework for people to better grasp the inner workings of OpsisDx.